

The next dialog shows all the matches that the stringApp finds for your disease query and selects the first one. The stringApp will retrieve a STRING network for the top-N proteins (by default 100) associated with the disease. In the import dialog, choose STRING: disease query as Data Source and type a disease of interest into the Enter disease term field (e.g. Go to the menu File → Import → Network from Public Databases. How is this network different from the protein-only network with respect to node types and the information provided in the Node Table? 1.3 Disease queries You can select the organism and number of additional interactors just like for the protein query above, and the disambiguation dialog also works the same way. In the import dialog, choose STITCH: protein/compound query as Data Source and type your favorite compound into the Enter protein or compound names or identifiers field (e.g. How many nodes are in the resulting network? How does this compare to the maximum number of interactors you specified? What types of information does the Node Table provide? 1.2 Compound queries Select the right one(s) you meant and continue by pressing the Import button. It lists all the matches that the stringApp finds for each ambiguous query term and selects the first one for each. Unless the name(s) you entered give unambiguous matches, a disambiguation dialog will be shown next. If you enter more than one protein name, the network will contain only the interactions among these proteins, unless you explicitly ask for additional proteins. By default, if you enter only one protein name, the resulting network will contain 10 additional interactors. The Maximum number of interactors determines how many interaction partners of your protein(s) of interest will be added to the network. You can select the appropriate organism by typing the name (e.g. In the import dialog, choose STRING: protein query as Data Source and type your favorite protein into the Enter protein names or identifiers field (e.g. In this exercise, we will perform some simple queries to retrieve molecular networks based on a protein, a small-molecule compound, a disease, and a topic in PubMed.

If you are not already familiar with the STRING and DISEASES databases, we highly recommend that you go through the short STRING exercises to learn about the underlying data before working with them in these exercises.
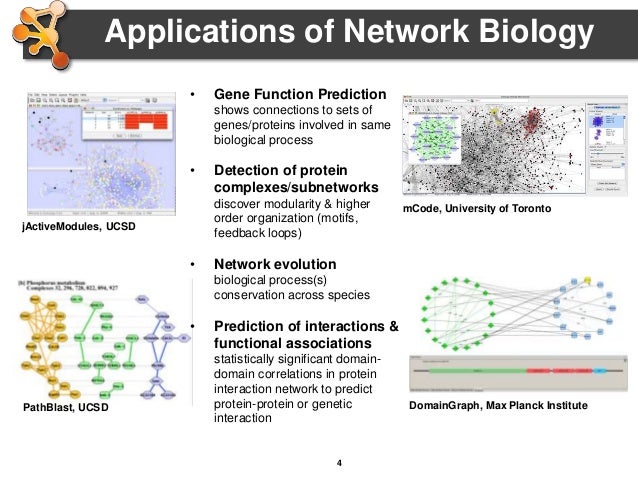
Similarly, make sure you have the yFiles Layout Algorithms, enhancedGraphics, and clusterMaker2 apps installed before closing the App Manager.
#CYTOSCAPE TUTORIAL INSTALL#
Search for the stringApp in the search field if it is not already installed, select it and press the Install button to install it. The exercises require you to have certain Cytoscape apps installed.
#CYTOSCAPE TUTORIAL UPDATE#
Then start Cytoscape and go to Apps → App Manager to check for new apps, install them and update the current ones if necessary. To follow the exercises, please make sure that you have the latest version of Cytoscape installed. identify functional modules through network clustering.perform enrichment analyses and visualize the results.import external data and map them onto a network.layout and visually style the resulting networks.retrieve networks for proteins of interest.In these exercises, we will use the stringApp for Cytoscape to retrieve molecular networks from the STRING database.


 0 kommentar(er)
0 kommentar(er)
